One shearing cell¶
The example in examples/oneCellShear presents a single red blood cell
(RBC) subjected to a shearing flow. The example provides a validation case for
the different material models used in HemoCell.
A single RBC is initialised in a fully periodic domain, i.e. periodicity is
enabled for all boundaries. The domain is then subjected to a shearing flow in
the x-direction. To achieve this flow field, the top and bottom surfaces are
subjected to velocity boundary conditions in opposing directions. The largest
diameter achieved by the RBC is reported and can be compared with respect to
experimental data.
After compilation, the example can be run using single core as:
# run the simulation from the `examples/oneCellShear` directory
mpirun -n 1 ./oneCellShear config.xml
# generate Paraview compatible output files
../../scripts/batchPostProcess.sh
The outcome files are generated in tmp/, where the flow field and particle
fields can be visualised separately by respectively viewing the
tmp/Fluid.*.xmf and tmp/RBC.*.xmf files.
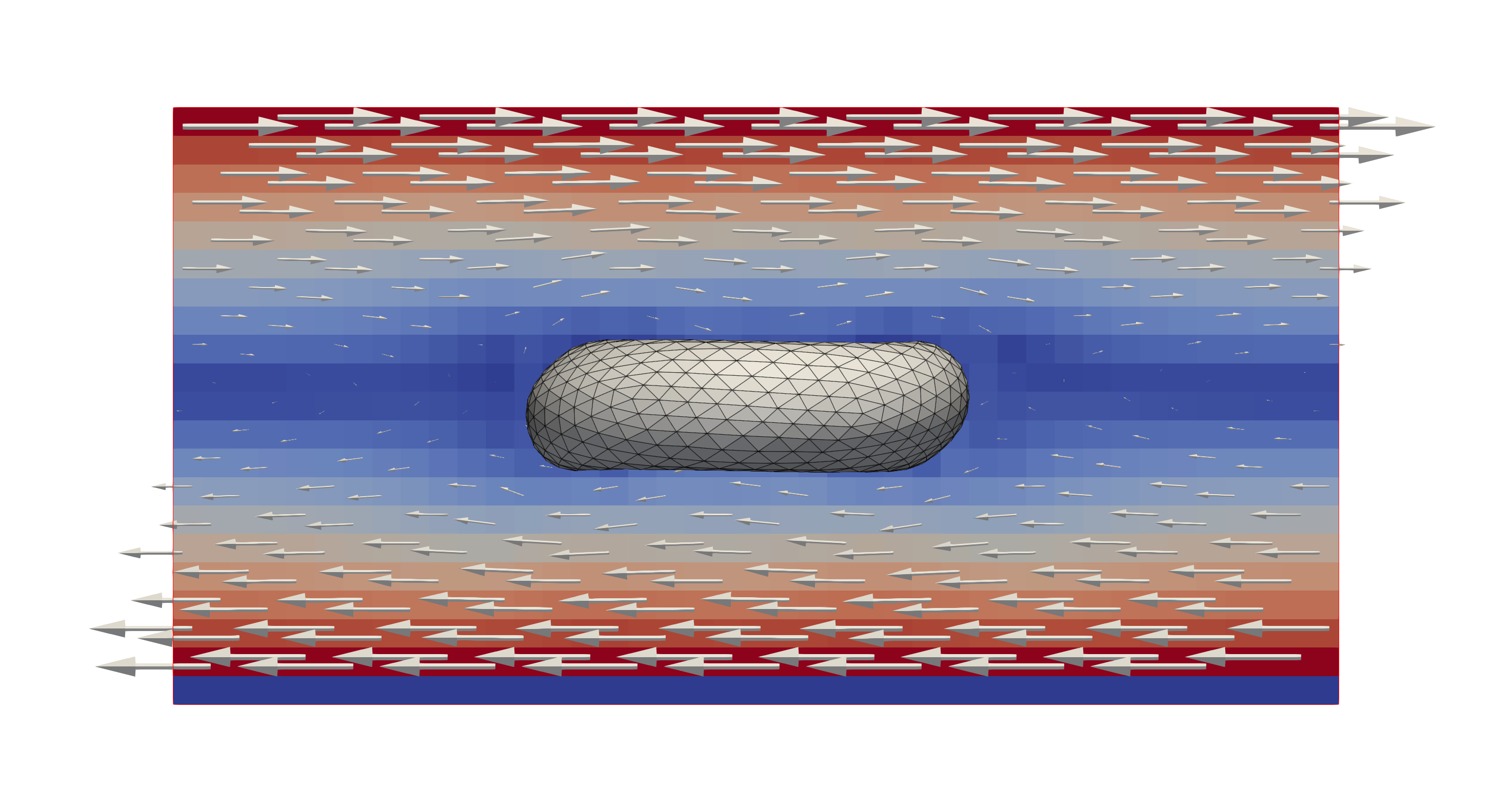
A single red blood cell subjected to a shearing flow in x-direction. The
colours and glyps indicate the flow velocity and direction. The effect of the
shearing flow becomes visible in the deformations of the RBC.¶
A gnuplot script is provided as well to visualise the largest diameter of the cell over the iterations, which can be invoked through
gnuplot shear.gpl
Tank treading¶
A simple variation of the single shearing RBC, is to consider the “tank
treading” case. This considers a problem where the RBC is rotated in the domain
and can be achieved by changing the loaded RBC properties by updating "RBC"
used in the oneCellShear.cpp example to "RBC_tt".
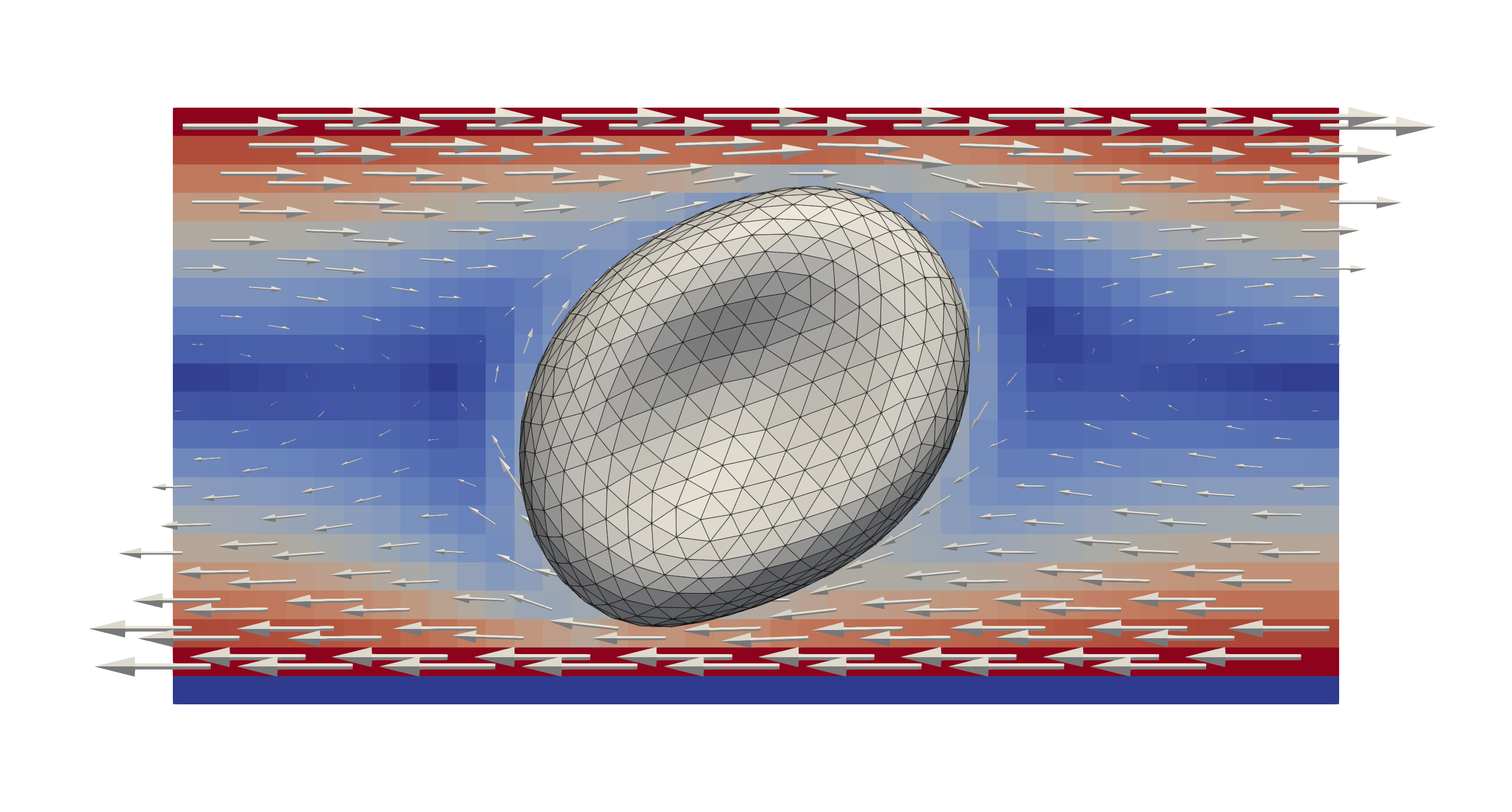
A single red blood cell subjected to a shearing flow in x-direction in
the “tank treading” configuration. The colours and glyps indicate the flow
velocity and direction. The cell now “rotates” along with the shearing flow
resulting in different deformations.¶
Configuration¶
The imposed velocity is derived from the shear rate provided in the configuration
file config.xml, where the shear rate can be set manually by updating the
<domain><shearrate> entry. Note, this value specifies the shear rate in
1/s.
Rendering¶
An example rendering pipeline with Blender is illustrated in the
hemocell/scripts/visualization/render_oneCellShear.py script, for more
information see Rendering with Blender.
An example rendering of the RBC using Blender.¶
